Actaea racemosa (root and rhizome)
(correct reference for Qiu et al) |
|||
| Line 165: | Line 165: | ||
The interpretation of NMR spectroscopic information for structure elucidation involves decoding of complex resonance patterns that contain valuable molecular information (δ and J), which is not readily accessible otherwise. We introduce a new concept of 2D-NMR barcoding that uses clusters of fingerprint signals and their spatial relationships in the δ−δ coordinate space to facilitate the chemical identification of complex mixtures. Similar to widely used general barcoding technology, the structural information of individual compounds is encoded as a specifics pattern of their C,H correlation signals. Software-based recognition of these patterns enables the structural identification of the compounds and their discrimination in mixtures. Using the triterpenes from various Actaea (syn. Cimicifuga) species as a test case, heteronuclear multiple-bond correlation (HMBC) barcodes were generated on the basis of their structural subtypes from a statistical investigation of their δH and δC data in the literature. These reference barcodes allowed in silico identification of known triterpenes in enriched fractions obtained from an extract of A. racemosa (black cohosh). After dereplication, a differential analysis of heteronuclear single-quantum correlation (HSQC) spectra even allowed for the discovery of a new triterpene. The 2D barcoding concept has potential application in a natural product discovery project, allowing for the rapid dereplication of known compounds and as a tool in the search for structural novelty within compound classes with established barcodes.<ref> | The interpretation of NMR spectroscopic information for structure elucidation involves decoding of complex resonance patterns that contain valuable molecular information (δ and J), which is not readily accessible otherwise. We introduce a new concept of 2D-NMR barcoding that uses clusters of fingerprint signals and their spatial relationships in the δ−δ coordinate space to facilitate the chemical identification of complex mixtures. Similar to widely used general barcoding technology, the structural information of individual compounds is encoded as a specifics pattern of their C,H correlation signals. Software-based recognition of these patterns enables the structural identification of the compounds and their discrimination in mixtures. Using the triterpenes from various Actaea (syn. Cimicifuga) species as a test case, heteronuclear multiple-bond correlation (HMBC) barcodes were generated on the basis of their structural subtypes from a statistical investigation of their δH and δC data in the literature. These reference barcodes allowed in silico identification of known triterpenes in enriched fractions obtained from an extract of A. racemosa (black cohosh). After dereplication, a differential analysis of heteronuclear single-quantum correlation (HSQC) spectra even allowed for the discovery of a new triterpene. The 2D barcoding concept has potential application in a natural product discovery project, allowing for the rapid dereplication of known compounds and as a tool in the search for structural novelty within compound classes with established barcodes.<ref> | ||
Qiu, F., McAlpine, J.B., Lankin, D.C., Burton, I., Karakach, T., Chen, S., Pauli G.F. 2014. 2D NMR Barcoding and Differential Analysis of Complex Mixtures for Chemical Identification: The Actaea Triterpenes ''Analytical Chemistry'' 86(8), 3964-3972. http://pubs.acs.org/doi/abs/10.1021/ac500188j</ref></blockquote> | Qiu, F., McAlpine, J.B., Lankin, D.C., Burton, I., Karakach, T., Chen, S., Pauli G.F. 2014. 2D NMR Barcoding and Differential Analysis of Complex Mixtures for Chemical Identification: The Actaea Triterpenes ''Analytical Chemistry'' 86(8), 3964-3972. http://pubs.acs.org/doi/abs/10.1021/ac500188j</ref></blockquote> | ||
| + | |||
| + | ===Cicek, ''et al.'', 2010=== | ||
| + | '''Development of a fast and convenient method for the isolation of triterpene saponins from Actaea racemosa by high-speed countercurrent chromatography coupled with evaporative light scattering detection.''' | ||
| + | |||
| + | <blockquote>'''Abstract.''' | ||
| + | |||
| + | In the present work, a fast and simple method for the separation and purification of triterpene saponins from Actaea racemosa was successfully established. Accelerated solvent extraction was used for defatting and extracting of the subaerial parts, giving a triterpene enriched crude extract. Size exclusion chromatography was used to separate actein and 23-epi-26-deoxyactein from other triterpenoids, which were collected in a third fraction. This most complex third fraction was applied to high-speed countercurrent chromatography, a well-established technique for the separation of saponins. Separation parameters were first optimized on an analytical level, using a hyphenated HSCCC-ELSD setup, before the system was scaled up to preparative size. The resulting two-phase solvent system, consisting of N-hexane-acetone-ethyl acetate-2-propanol-ethanol-water (3.5 : 1 : 2 : 1 : 0.5 : 2, v/v/v/v/v/v), enabled the isolation of 23-O-acetylshengmanol-3-O- beta-D-xylopyranoside (17.4 mg), cimiracemoside D (19.5 mg), 25-O-acetylcimigenol-3-O-beta-D-xylopyranoside (7.1 mg) and the aglycone cimigenol (5.9 mg). Purity of the isolated substances was 96.8 %, 96.2 %, 97.9 %, and 98.4 %, respectively. The same method was suitable for the purification of actein and 23-epi-26-deoxyactein, with purities of 97.0 % and 98.3 %.<ref> | ||
| + | Cicek, S.S., Schwaiger, S., Ellmerer, E.P., Stuppner, H. 2010. Development of a fast and convenient method for the isolation of triterpene saponins from Actaea racemosa by high-speed countercurrent chromatography coupled with evaporative light scattering detection. ''Planta Med.'' 76(5):467-73. http://www.ncbi.nlm.nih.gov/pubmed/19847744</ref></blockquote> | ||
=Sources= | =Sources= | ||
<references /> | <references /> | ||
Revision as of 19:22, 17 April 2014
Contents |
Nomenclature
Actaea racemosa L. Ranunculaceae
Syn. Cimicifuga racemosa (L.) Nutt.
Standardized common name (English): black cohosh
Botanical Voucher Specimen
|
|
Organoleptic Characteristics
|
Macroscopic Characteristics
|
Microscopic Characteristics
|
High Performance Thin Layer Chromatographic Identification
|
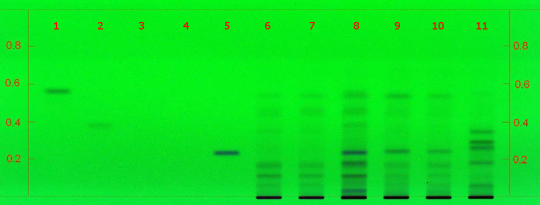
Black cohosh (root) (Actaea racemosa) Lane Assignments Lanes, from left to right (Track, Volume, Sample):
Reference Sample(s) Reference: Dissolve 1mg of actein, 23-epi-26-deoxyactein, isoferulic acid, cimifugin and/or norcimifugin individually in 10 mL of methanol. Stationary Phase Stationary phase, i.e. Silica gel 60, F254 Mobile Phase Toluene, ethyl formate, formic acid 50:30:20 (v/v/v) Sample Preparation Method Sample: Mix 0.5 g of powdered sample with 10 mL of an ethanolwater mixture (1:1) and sonicate for 10 minutes, then centrifuge or filter the solutions and use the supernatants/filtrates as test solutions. Sulfuric Acid Reagent: Preparation: 20 mL of sulfuric acid are mixed with 180 mL of ice-cooled methanol Detection Method UV/Vis, Sulfuric Acid Reagent Use: Dip (time 0, speed 5), heat at 100°C for 5 min Other Notes Compare result under UV 254 nm and white RT with reference images in Image Comparison Viewer. The fingerprint of the test solution is similar to that of the corresponding botanical reference sample. Additional weak zones may be present. C. racemosa Identification: Under UV 254 nm the chromatogram obtained with the test solution does not show a dark zone at the position of Cimifugin in the reference solution or just above it. Under white RT a characteristic fingerprint is detected. Below Cimifugin there are two violet zones. Just above the application position two brown zones are detected (blue arrow). There is no brown zone at the position of Cimifugin, but there might be a faint violet zone. There is a brown zone at the position of actein as well as two brown zone just below. Just above the actein zone is a very thin brown zone (green arrow). Further methodology available here, in Identification of black cohosh by HPTLC. Source: AHPA Practical, CAMAG HPTLC [7] |
Supplementary Information
Black cohosh (root) (Actaea racemosa, syn Cimicifuga racemosa)
General Characteristics AHPA recommends in its Known Adulterants list that appropriate steps be taken to assure that this raw material is free of the noted adulterant. Contact AHPA for additional information regarding relevant analytical methods or follow this link for more information.
Reported Adulterants Chinese cimicifuga (root) (Actaea spp.). Also known as sheng ma or Rhizoma Cimicifugae; consists of Actaea cimicifuga, syn. Cimicifuga foetida; Actaea dahurica, syn. C. dahurica; A. heracleifolia, syn. C. heracleifolia; and possibly other Asian species of Actaea.
Source: AHPA Known Adulterants [8]
AHPA Practical: Detecting Adulteration of Black cohosh root/rhizome
Reproduced from [1].
Introduction
The American Herbal Products Association is providing here analytical tools and methods to identify adulteration of raw materials and dietary ingredients labeled as black cohosh (Actaea racemosa syn. Cimicifuga racemosa) root/rhizome, or extracts thereof. This information is provided for industry in order to deal appropriately with reports of adulteration of this ingredient and make wise purchasing decisions.
Background
The economic adulteration of black cohosh root and rhizome with other species is well established. AHPA’s guidance policy on Known Adulterants identifies Chinese cimicifuga root/rhizome, also known as sheng ma or Rhizoma Cimicifugae as known black cohosh adulterants. This material commonly consists of Actaea cimicifuga, syn. Cimicifuga foetida; Actaea dahurica, syn. C. dahurica; A. heracleifolia, syn. C. heracleifolia; and possibly other Asian species of Actaea. The citation below provides a review of Rhizoma Cimicifugae for those wanting additional information.
Li JX, Yu ZY. Cimicifugae rhizoma: from origins, bioactive constituents to clinical outcomes. Curr Med Chem. 2006;13(24):2927-51.
The black cohosh monograph in the Natural Health Ingredients Database of the Canadian Natural Health Products Directorate at Health Canada, available online here or as a pdf file here, references instances where adverse events associated with ingestion of products labeled as containing black cohosh (Actaea racemosa syn. Cimicifuga racemosa) were found to contain plant species that were not black cohosh upon laboratory analysis. The Canadian Adverse Reaction Newsletter (2010 Jan;20(1):1-2) provided specifics on this situation as covered in a January 14, 2010 AHPA Update.
Analytical tools
There are several analytical tools available for the authentication of genuine black cohosh and its differentiation from other species. Two papers detailing the use of high performance thin-layer chromatography (HPTLC) are cited below. The first focuses on detection of black cohosh adulteration by other North American Actaea species while the second deals more specifically with differentiation of black cohosh from Chinese cimicifuga. The full methodology for the HPTLC analysis of the second citation can be accessed via this: Identification of black cohosh by HPTLC.
The third citation below employed phytochemical “fingerprinting” using high-performance liquid chromatography with a photo-diode array detector (HPLC-PDA) and liquid chromatography with mass spectrometry detection (LC-MS) to authenticate black cohosh and differentiate it from 14 other Actaea species.
Verbitski SM, Gourdin GT, Ikenouye LM, McChesney JD, Hildreth J. Detection of Actaea racemosa adulteration by thin-layer chromatography and combined thin-layer chromatography-bioluminescence. J AOAC Int. 2008 Mar-Apr;91(2):268-75.
Ankli A, Reich E, Steiner M. Rapid high-performance thin-layer chromatographic method for detection of 5% adulteration of black cohosh with Cimicifuga foetida, C. heracleifolia, C. dahurica, or C. americana. J AOAC Int. 2008 Nov-Dec;91(6):1257-64.
Jiang B, Ma C, Motley T, Kronenberg F, Kennelly EJ. Phytochemical fingerprinting to thwart black cohosh adulteration: a 15 Actaea species analysis. Phytochem Anal. 2011 Feb 19. doi: 10.1002/pca.1285. [Epub ahead of print] Supporting information is available here.
The black cohosh monograph in the Natural Health Ingredients Database of the Canadian Natural Health Products Directorate at Health Canada, available online here or as a pdf file here, references instances where adverse events associated with ingestion of products labeled as containing black cohosh (Actaea racemosa syn. Cimicifuga racemosa) were found to contain plant species that were not black cohosh upon laboratory analysis. The Canadian Adverse Reaction Newsletter (2010 Jan;20(1):1-2) provided specifics on this situation as covered in a January 14, 2010 AHPA Update.
The American Herbal Pharmacopoeia and the USP Dietary Supplements Compendium also contain useful information for the differentiation of black cohosh and other species representing common black cohosh adulterants. Authenticated black cohosh reference materials are available from both organizations as well as AHPA member companies Alkemists Laboratories, Botanical Liaisons, and Chromadex. DNA testing can be used to verify authentic species, and rule out the presence of common or other unexpected DNA-containing adulterants or contaminants. Work in this area is currently being conducted by AHPA member AuthenTechnologies.
Other Publications
Qiu, et al., 2014
2D NMR Barcoding and Differential Analysis of Complex Mixtures for Chemical Identification: The Actaea Triterpenes,
Abstract. The interpretation of NMR spectroscopic information for structure elucidation involves decoding of complex resonance patterns that contain valuable molecular information (δ and J), which is not readily accessible otherwise. We introduce a new concept of 2D-NMR barcoding that uses clusters of fingerprint signals and their spatial relationships in the δ−δ coordinate space to facilitate the chemical identification of complex mixtures. Similar to widely used general barcoding technology, the structural information of individual compounds is encoded as a specifics pattern of their C,H correlation signals. Software-based recognition of these patterns enables the structural identification of the compounds and their discrimination in mixtures. Using the triterpenes from various Actaea (syn. Cimicifuga) species as a test case, heteronuclear multiple-bond correlation (HMBC) barcodes were generated on the basis of their structural subtypes from a statistical investigation of their δH and δC data in the literature. These reference barcodes allowed in silico identification of known triterpenes in enriched fractions obtained from an extract of A. racemosa (black cohosh). After dereplication, a differential analysis of heteronuclear single-quantum correlation (HSQC) spectra even allowed for the discovery of a new triterpene. The 2D barcoding concept has potential application in a natural product discovery project, allowing for the rapid dereplication of known compounds and as a tool in the search for structural novelty within compound classes with established barcodes.[9]
Cicek, et al., 2010
Development of a fast and convenient method for the isolation of triterpene saponins from Actaea racemosa by high-speed countercurrent chromatography coupled with evaporative light scattering detection.
Abstract. In the present work, a fast and simple method for the separation and purification of triterpene saponins from Actaea racemosa was successfully established. Accelerated solvent extraction was used for defatting and extracting of the subaerial parts, giving a triterpene enriched crude extract. Size exclusion chromatography was used to separate actein and 23-epi-26-deoxyactein from other triterpenoids, which were collected in a third fraction. This most complex third fraction was applied to high-speed countercurrent chromatography, a well-established technique for the separation of saponins. Separation parameters were first optimized on an analytical level, using a hyphenated HSCCC-ELSD setup, before the system was scaled up to preparative size. The resulting two-phase solvent system, consisting of N-hexane-acetone-ethyl acetate-2-propanol-ethanol-water (3.5 : 1 : 2 : 1 : 0.5 : 2, v/v/v/v/v/v), enabled the isolation of 23-O-acetylshengmanol-3-O- beta-D-xylopyranoside (17.4 mg), cimiracemoside D (19.5 mg), 25-O-acetylcimigenol-3-O-beta-D-xylopyranoside (7.1 mg) and the aglycone cimigenol (5.9 mg). Purity of the isolated substances was 96.8 %, 96.2 %, 97.9 %, and 98.4 %, respectively. The same method was suitable for the purification of actein and 23-epi-26-deoxyactein, with purities of 97.0 % and 98.3 %.[10]
Sources
- ↑ Tropicos.org. Missouri Botanical Garden. 05 Aug 2013 http://www.tropicos.org/Image/100105518
- ↑ United States Dispensatory (1918)
- ↑ United States Dispensatory (1918)
- ↑ Alkemist Laboratories http://www.alkemist.com
- ↑ United States Dispensatory (1918)
- ↑ Elan M. Sudberg, Alkemist Laboratories http://www.alkemist.com
- ↑ AHPA Practical, CAMAG HPTLC http://www.camag.com/index.php
- ↑ AHPA Known Adulterants http://www.ahpa.org/
- ↑ Qiu, F., McAlpine, J.B., Lankin, D.C., Burton, I., Karakach, T., Chen, S., Pauli G.F. 2014. 2D NMR Barcoding and Differential Analysis of Complex Mixtures for Chemical Identification: The Actaea Triterpenes Analytical Chemistry 86(8), 3964-3972. http://pubs.acs.org/doi/abs/10.1021/ac500188j
- ↑ Cicek, S.S., Schwaiger, S., Ellmerer, E.P., Stuppner, H. 2010. Development of a fast and convenient method for the isolation of triterpene saponins from Actaea racemosa by high-speed countercurrent chromatography coupled with evaporative light scattering detection. Planta Med. 76(5):467-73. http://www.ncbi.nlm.nih.gov/pubmed/19847744